New techniques in medical informatics lead to improved diagnosis of MDS
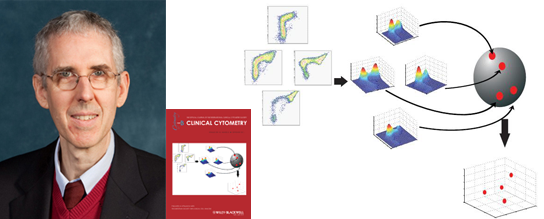
The technique involves a visualization method that renders clinical flow cytometry data more interpretable to pathologists.
 Enlarge
Enlarge
Research by Prof. Al Hero and colleagues in the area of medical informatics is leading to better diagnosis and prognosis of patients with serious blood borne diseases. The researchers have recently improved upon a technique they developed that allows doctors to better diagnose a condition that often leads to anemia and in acute cases, leukemia.
The technique involves a visualization method, called Fisher Information Non-parametric Embedding (FINE), that renders clinical flow cytometry data more interpretable to pathologists. FINE measures information distances among flow cytometry datasets using principles of manifold learning and information geometry to cluster the high dimensional cytometric data.
This research was first reported in the paper “Analysis of clinical flow cytometric immunophenotyping data by clustering on statistical manifolds: Treating flow cytometry data as high-dimensional objects” by William G. Finn, Kevin M. Carter, Raviv Raich, Lloyd M. Stoolman, and Al O. Hero. The paper was selected as the Best Original Paper published in Clinical Cytometry for 2008-2009 (originally published in Vol. 76B 1, 107, 2009). [more info]
In a new paper, “Immunophenotypic signatures of benign and dysplastic granulopoiesis by cytomic profiling,” by William G. Finn, Alexandra M. Harrington, Kevin M. Carter, Raviv Raich, Steven H. Kroft and Alfred O. Hero III, Cytometry Part B: Clinical Cytometry (Sept. 2011), the authors applied FINE and a novel visualization method called Information Preserving Components Analysis (IPCA) to the particularly difficult problem of diagnosing myelodysplastic syndromes (MDS). MDS are a group of diseases of the blood and bone marrow that can lead to anemia or in the worst case, leukemia.
The paper demonstrates that by using FINE and IPCA pathologists can better separate malignant from benign MDS in a cohort of 37 patients tested.
The clinical study was led by U-M pathologist Dr. William Finn. Both FINE and IPCA were developed by Prof. Hero, his former PhD student Kevin Carter (now at MIT Lincoln Laboratory), and his former post-doctoral student Raviv Raich (now at Oregon State University). FINE was described above; IPCA is a new method for visualizing data in a way that preserves information content in a precise mathematical way. A tutorial for FINE and IPCA was recently published in the paper, “Information-Geometric Dimensionality Reduction” IEEE Signal Processing Magazine, vol. 28, no. 2, pp. 89-99, Mar. 2011.
*****************************
Prof. Hero is a member of the Systems Laboratory in the EECS Department, division of Electrical and Computer Engineering. His research interests include inference in sensor networks, adaptive sensing, bioinformatics, inverse problems, and statistical signal and image processing. He holds joint appointments in the Department of Biomedical Engineering and the Department of Statistics, and during the summer holds an appointment as Digiteo Chaire d’Excellence at the Digiteo Research Park in Information Science and Technology, Paris, France. He is also affiliated with the U-M Program in Biomedical Science (PIBS) and the U-M Graduate Program in Applied and Interdisciplinary Mathematics (AIM).
 MENU
MENU 
